|
I'm a researcher based in San Francisco, CA. My interests include applications of machine learning to problems in health and genomics. Previously, I was at Google Research where I worked on genomics and multimodal machine learning. I completed my Masters and undergraduate studies at UC Berkeley, where I was a part of the RISELab. At Google, I developed DeepConsensus for error correction in PacBio sequencing data and worked on DeepVariant for small variant calling. My work in multimodal ML includes text-to-image generation with Parti and Multisearch for Google Lens. Email / Google Scholar / Twitter / GitHub |

|
|
|
|
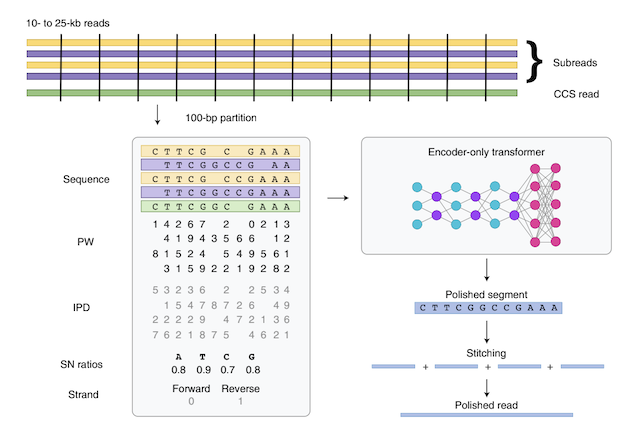
Nature Biotechnology, 2022 Paper / Blog Post / GitHub DeepConsensus uses a Transformer to reduce errors in PacBio Circular Consensus Sequencing (CCS) data by 59%. This method is now deployed on PacBio Revio sequencers. Awarded Best Scientific Breakthrough (one project across Alphabet). |

|
TMLR, 2022 Paper / Website / Blog Post / GitHub Parti (Pathways Autoregressive Text-to-Image) is an autoregressive text-to-image generation model that achieves high-fidelity photorealistic image generation and supports content-rich synthesis involving complex compositions and world knowledge. |
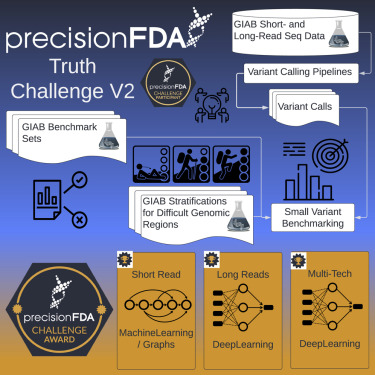 |
Cell Genomics, 2022 Paper / Challenge Results / Blog Post This challenge assessed variant calling pipelines on reference datasets, with a focus on difficult-to-map regions, segmental duplications, and the Major Histocompatibility Complex (MHC). DeepVariant recevied best overall accuracy in 3 out of 4 sequencing instrument categories, and we released an improved version of the submitted model as DeepVariant v1.0. |

|
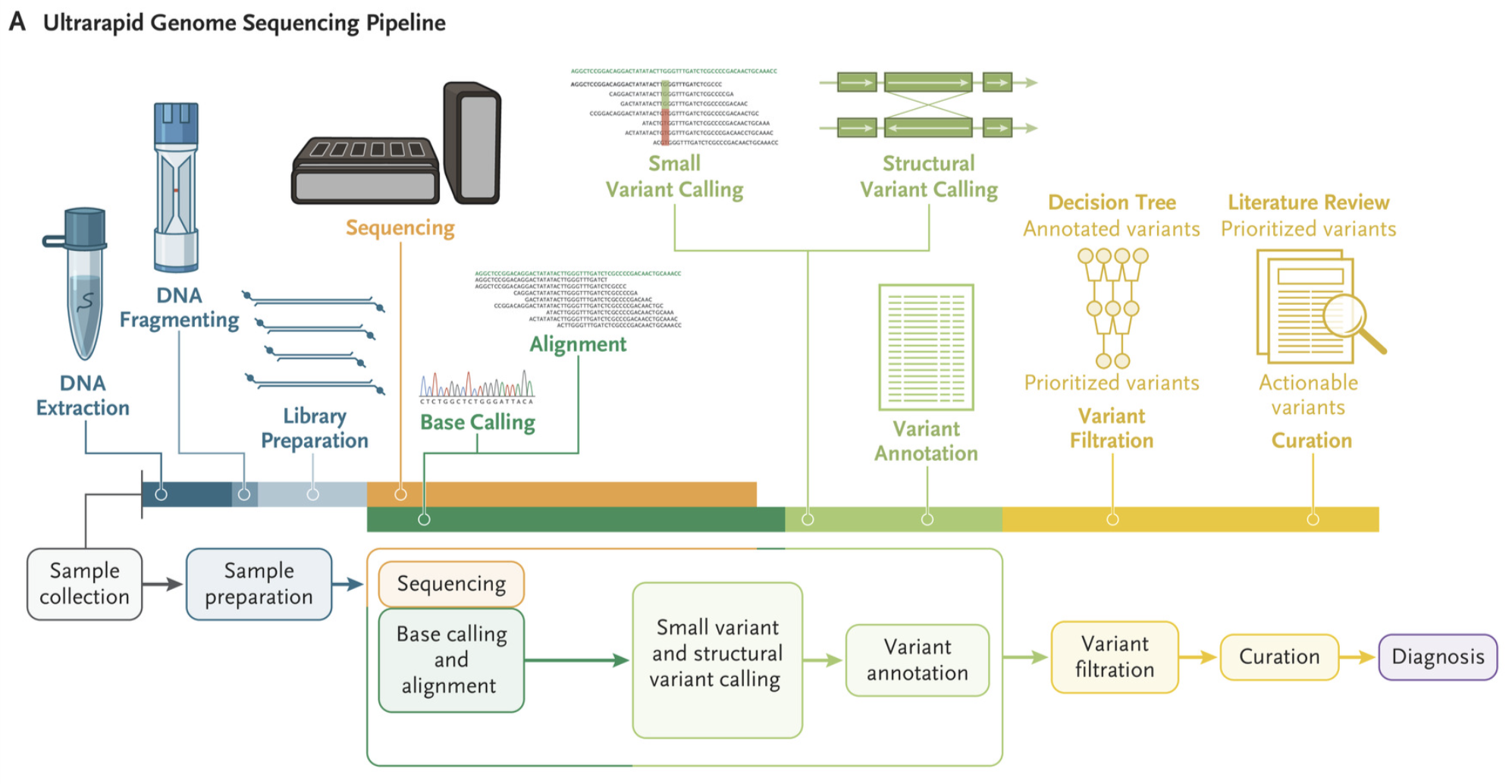
Nature Biotechnology, 2022 Paper / Blog Post This pipeline performs distributed whole-genome Nanopore sequencing, near real-time base calling and alignment, accelerated variant calling, and custom variant prioritization. We received a Guiness World Record for fastest DNA sequencing technique. |
|
New England Journal of Medicine, 2022 Paper Whole-genome sequencing was performed for twelve patients in critical care using an accelerated pipeline. The shortest time from arrival of the blood sample to the initial diagnosis was 7 hours 18 minutes. A pathogenic or likely pathogenic variant was identified in five out of 12 patients. |
|
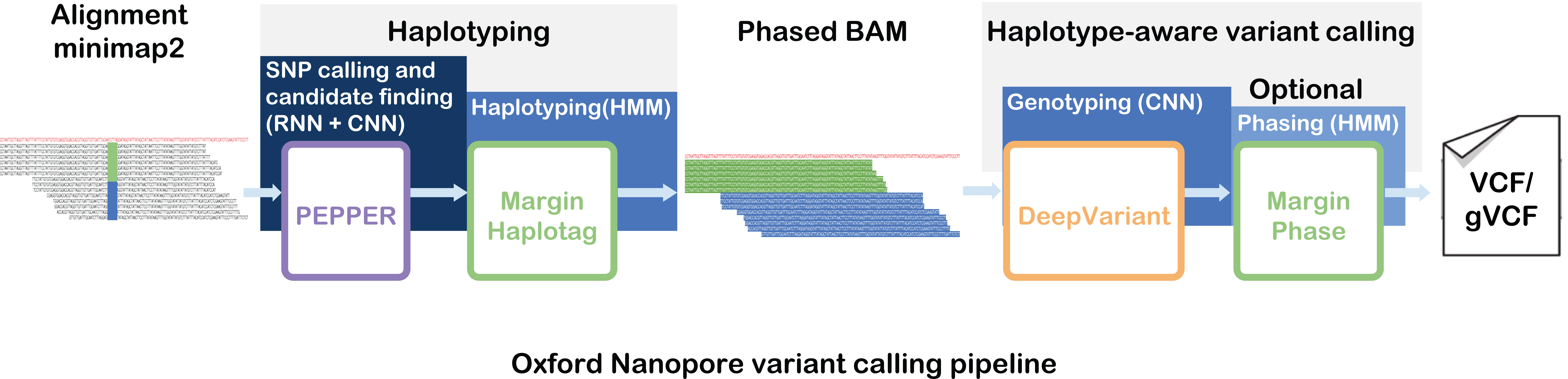
Nature Methods, 2021 Paper / GitHub PEPPER-Margin-DeepVariant is a haplotype-aware variant calling pipeline for Oxford Nanopore data that produces state-of-the-art results. |
|
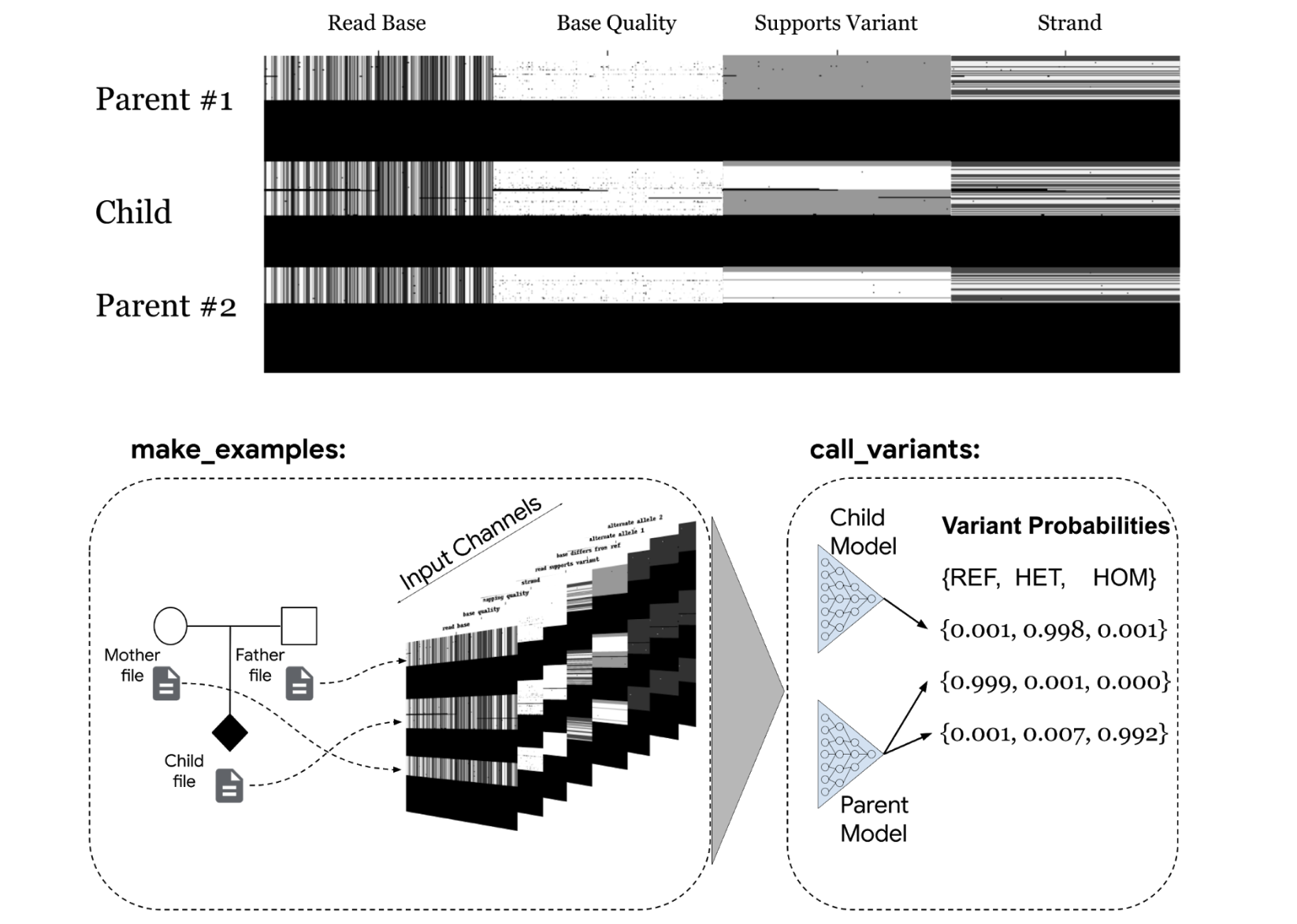
bioRxiv, 2021 Paper / Blog Post We extend DeepVariant to to consider the genetic variants in mother-father-child trios. Coexamination of available parental data improves the accuracy of variant calls for the child. The DeepTrio model was released as part of DeepVariant v1.1. |
|
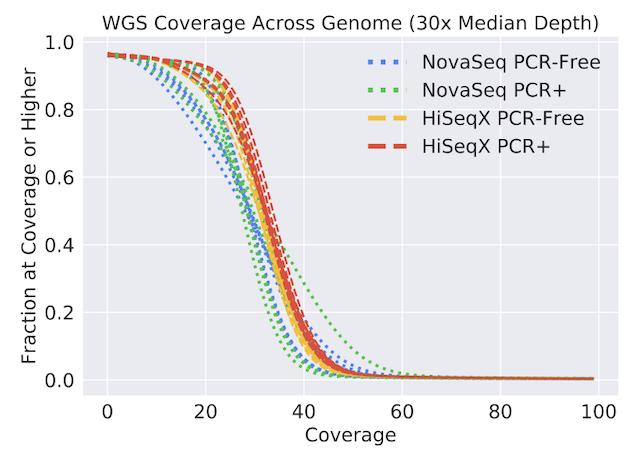
bioRxiv, 2020 Paper / Dataset We provide a benchmark dataset of WGS and WES samples from HG001-7 and NA12878/9 from a variety of sequencers. We characterize properties of the data and present variant calling results on these samples. |
|
Design and source code from Jon Barron's website. |